Signal Peptide Website: An Information Platform for Signal Sequences and Signal Peptides
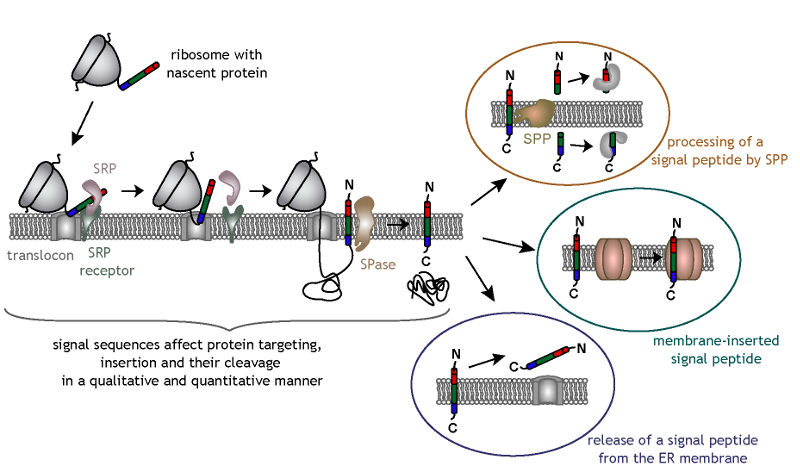
N-terminal signal sequences mediate targeting of nascent secretory and membrane proteins to the endoplasmic reticulum (ER) in a signal
recognition particle (SRP)-dependent manner. Signal sequences have a tripartite structure, consisting of a hydrophobic core region (h-region)
flanked by an n- and c-region. The latter contains the signal peptidase (SPase) consensus cleavage site. Usually, signal sequences are cleaved
off co-translationally so that signal peptides and mature proteins are generated. Signal sequences are extremely variable in length and amino
acid composition. This variability suggests that ER targeting and the steps beyond like protein insertion and SPase cleavage are affected by
the signal sequence.
Signal sequence variability may account for additional so called post-targeting functions of signal peptides. Interestingly, some signal
peptides are further processed by an intramembrane cleaving protease named signal peptide peptidase (SPP), and the resulting N-terminal signal
peptide fragments are released into the cytosol. Alternatively, signal peptides remain membrane-inserted and can be part of a protein complex,
while other signal peptides are released as such from the ER membrane. These signal peptides or signal peptide fragments are known to have
diverse functions, either together with or independent of their corresponding mature proteins.
References:
1.) von Heijne G. (1983) Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem 133 (1) 17-21
2.) Martoglio B. and Dobberstein B. (1998) Signal sequences: More than just greasy peptides. Trends Cell Biol 8 (10), 410-5
3.) Hegde R.S. and Bernstein H.D. (2006) The surprising complexity of signal sequences. Trends Biochem Sci 31(10), 563-71
4.) Kapp K., Schrempf S., Lemberg M.K. and Dobberstein B. (2009) Post-Targeting Functions of Signal Peptides. Chapter in: Protein Transport into the Endoplasmic Reticulum, Landes Bioscience